

Projects in progress:
Development of a computationally efficient model of the human heart
Three-dimensional organization of ventricular fibrillation in human heart

Recent presentations:
Human ventricular cell (TNNP) model
Recently we introduce a mathematical model of the action potential of
human ventricular cells that, while including a high level of
electrophysiological detail, is computationally cost-effective enough
to be applied in large-scale spatial simulations for the study of
reentrant arrhythmias (see figure below). The model is based on recent
experimental data on most of the major ionic currents and reproduces
properties of different cell types (endocardial, epicardial and M
cells), the experimentally observed data on action potential duration
restitution and the conduction velocity restitution. We use this model
to study dynamics of spiral wave rotation in 2D and in anatomically
accurate model of human heart.
( New version: K.H.W.J. ten Tusscher and A.V. Panfilov, 2006, American Journal of Physiology, 291(3):H1088-100. (Full text (HTML) and PDF for AJP subscribers); PubMed ID: 16565318)
Poster, for HRS 2009 meeting in Boston

( K.H.W.J. ten Tusscher, D. Noble, P.J. Noble, and A.V. Panfilov, 2004, American Journal of Physiology, 286, H1573-H1589. (Full text (HTML) and PDF for AJP subscribers); PubMed ID: 14656705)
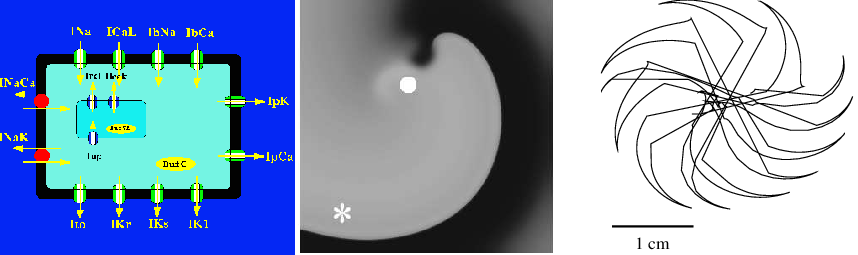
Schematic representation(left); Spiral wave dynamics (middle) Core of spiral wave (left)
Links
The webpage of K. Ten Tusscher, including PhD thesis
The source codes

Anatomically accurate modelling



 HUMAN VENTRICULAR MODEL together with Olivier Bernus and Kirsten Ten Tusscher
HUMAN VENTRICULAR MODEL together with Olivier Bernus and Kirsten Ten Tusscher







Reactio-diffusion-contraction systems (work in progress with
M.Nash )




Nash MP, Panfilov AV. : Prog Biophys Mol Biol. 2004 Jun-Jul;85(2-3):501-22.
Panfilov AV, Keldermann RH, Nash MP.:Phys Rev Lett. 2005 Dec 16;95(25):258104
Panfilov AV, Keldermann RH, Nash MP.:Proc. Natl. Acad. Sci. USA, 2007, v.104,p.7922-7926